|
|
What is PathJam?
PathJam is a public tool which provides an intuitive and user-friendly
framework for biological pathway analysis of human gene lists. This server
integrates pathway-related annotations from several public sources
(Reactome, KEGG, Biocarta, etc) making easier the understanding of gene
lists of interest.
Using PathJam you will obtain interactive graphs linking genes and pathway
annotations, exportable tables (.txt, .xls) including the annotations for
your lists and useful plots showing the connectivity, in terms of pathways
activity, for the genes.
PathJam has been thought to produce compatible outputs with well-established
analysis methods (i.e. GSEA .gmx format) and can be used as a widget in
CARGO server.
Currently,
genes are indexed from
pathways, available from
The Pathjam's index was updated on: Wed Oct 13 10:24:00 CEST 2010
Requirements
PathJam has been developed under Java techonology. This application is distributed as a Java applet inside a web page.
So, in order to be able to execute PathJam, your browser should support Java 1.5 or later.

At this point, IE Explorer (6 and 7), Mozilla Firefox (2 and 3) and Apple Safari are known to work.
Acknowledgements
PathJam displays information from different sources. We are thankful to the developers and Institutions for providing freely available services:
|
Quick start
To understand how PathJam works, in the following steps we
have a fast look at some of the available funcionalities:
1. Search for a single gene or list
of gene identifiers. The following gene identifiers can be used
for searching purpouses: Gene Symbol, Entrez Gene, Ensembl,
SwissProt and OMIM.
2. Once PathJam has executed your query,
search results are inmediately showed in a Java Applet.
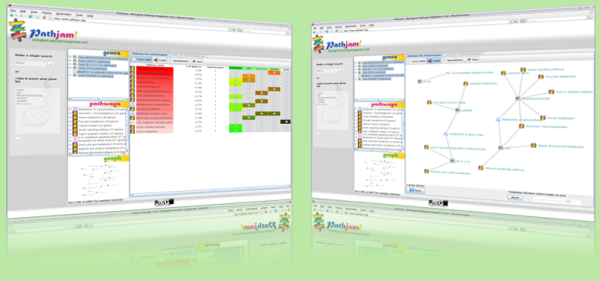
3. Click on one of the genes (or a group of
them) from the list and the tab Pathways for selected genes
will show you the pathways where each of selected genes are
involved.
4. Click on one of the pathways (or a group
of them) from the list and the tab Genes for selected pathways
will show you the genes which each of selected pathways contains.
5. In both cases, different views showing the relations
between genes and pathways become available: graph view, tabular
view and cross table view. In addition, you can save the current view as image (if graph view is selected) or as cvs file (if tabular view is selected).
Feedback
Please, give us your feedback here!
|
|



 for developers
for developers in collaboration with
in collaboration with 


